Modeling Controls¶
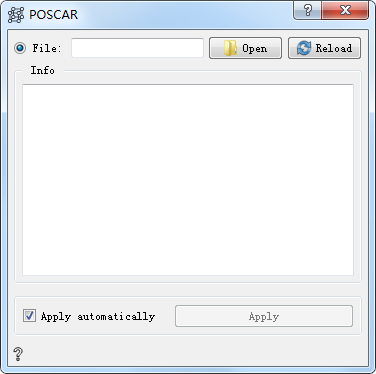
POSCAR Controls¶
Read files in POSCAR、cif、xsf and xyz formats.
Input |
Output |
|---|---|
None |
Structure: pymatgen class |
Parameter Configuration:mainly used to load and display atomic structure information, double-click the icon to open the parameter configuration.
Open Open and load the file
Reload Reload file
Info Show atomic structure information
Apply automatically If checked, the output of the control can be automatically passed to the next control. If unchecked, you can manually click Apply for transmission, which is checked by default.
Builder Controls¶
Complex models such as grain boundary, doping and vacancy can be constructed.Various methods such as finding grain boundaries, constructing grain boundaries, Interstitial doping, vacancy doping, substitution atom doping and arbitrary location doping are supported.
Input |
Output |
|
|---|---|---|
Structure |
Structure |
Build Structures |
Note
The following parameters configuration, with * is required, and with no * is optional.
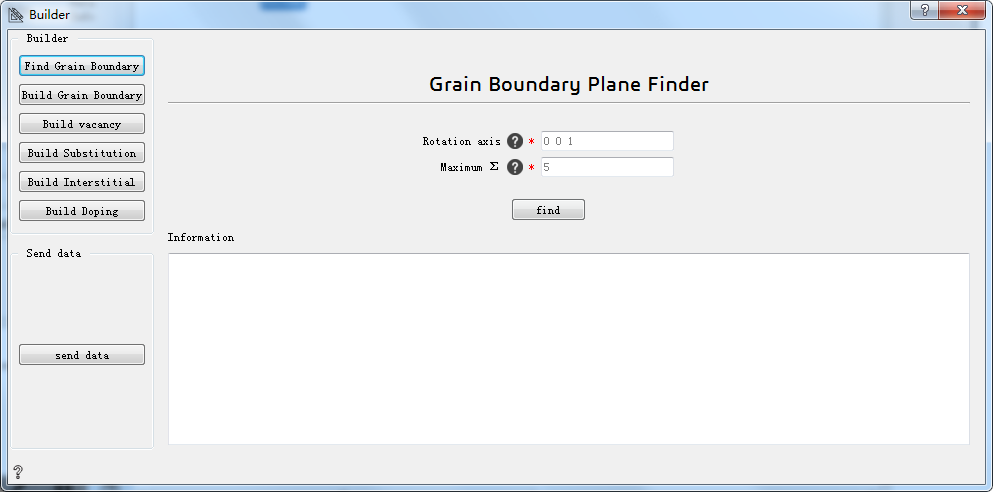
4.1.2.1 Find Grain Boundary:find grain boundary¶
Rotation axis* Specify the axis of rotation between two grains, such as 0 0 1
Maximum Σ* Maximum fitting degrees of freedom
find Click to find the specified boundary conditions
Information Show grain boundary structure information
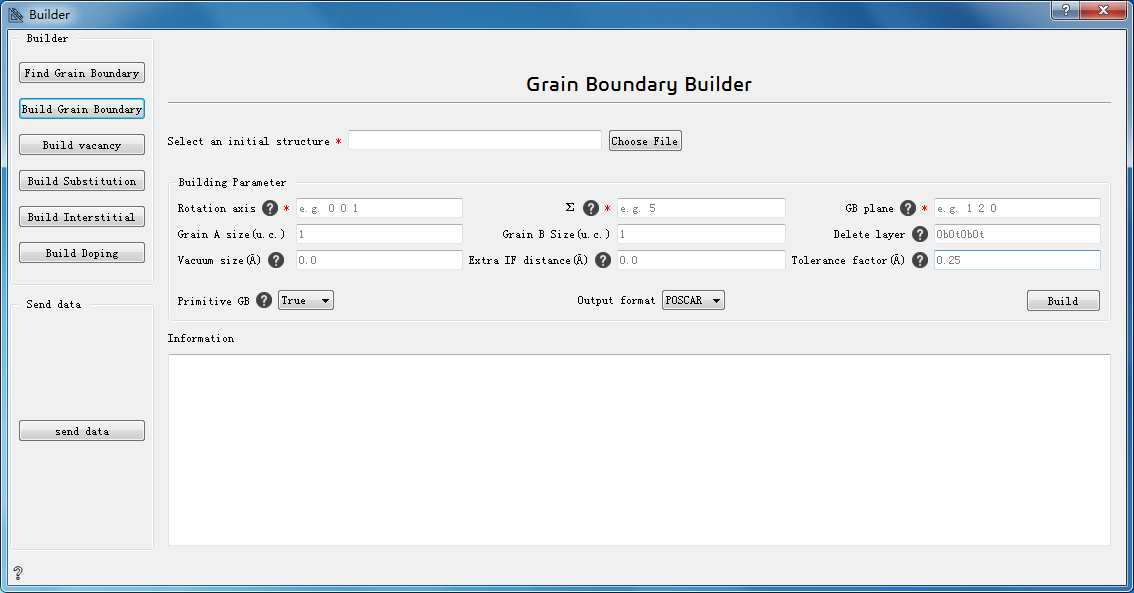
Build Grain Boundary:build grain boundary¶
Choose File* Choose the initial structure
Rotation axis* The common rotation axis of two grains, such as 0 0 1
Σ* Fit the degree of freedom, such as 5
GB plane* Grain boundary orientation, such as 1 2 0
Grain A size(u. c.) The size of grain A
Grain B size(u. c.) The size of grain B
Delete layer Delete the surface layer on either side of each grain. 8 characters must be input. The first four characters are for grain A and the last four characters are for grain B. ‘b’ for the bottom and ‘t’ for the top. The integer represents the number of layers to remove. The default is:0b0t0b0t,which means there are no layers to remove.
Vacuum size(Å) Set the thickness of the vacuum layer at the grain boundary (for grain boundaries with plate models)
Extra IF distance(Å) Add an extra distance between the interfaces of the two grains
Tolerance factor(Å) A tolerance factor that determines whether two atoms are in the same plane
primitive GB Whether to get the primitive structure of grain boundary
Output format Choose output format
Build Build grain boundary
Information Show the constructed grain boundary structure information
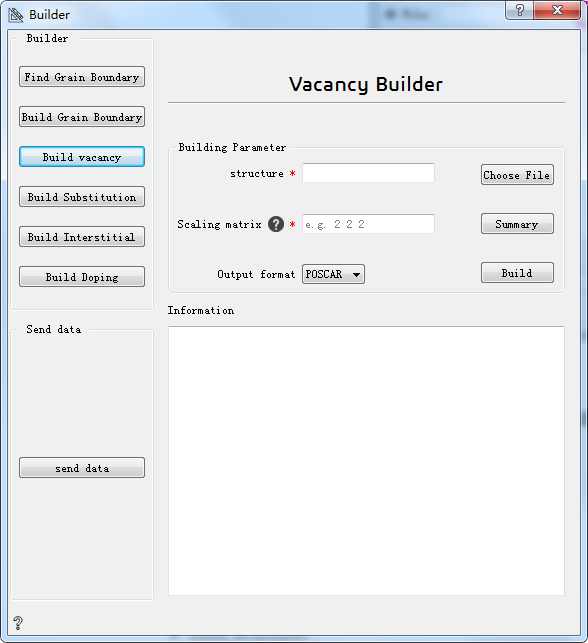
Build vacancy:build vacancy¶
Choose File* Choose the initial structure
Scaling matrix* Scaling matrix used to build supercells. The options include: one integer, representing expand multiple; three vectors,representing three directions at the same time to enlarge the corresponding multiples; nine matrices,representing to enlarge corresponding multiple in matrix.
Summary Offer all the information necessary to build a vacancy
Output format Choose output format
Build Build vacancy
Information Show vacancy structure information
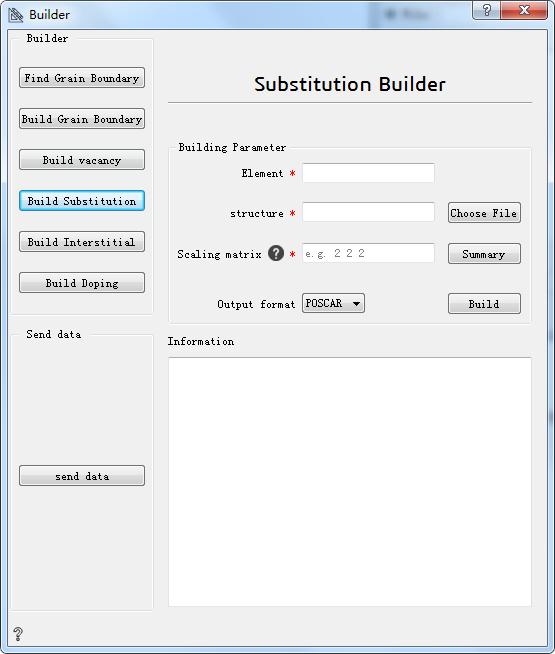
Build Substitution:build substitution atom¶
Element* Choose the element to substitute
Choose File* Choose the initial structure
Scaling matrix* Scaling matrix used to build supercells. The options include: one integer, representing expand multiple; three vectors,representing three directions at the same time to enlarge the corresponding multiples; nine matrices,representing to enlarge corresponding multiple in matrix.
Summary Offer all the information necessary to build substitution structure
Output format Choose output format
Build Build substitution
Information Show information about the substitution structure
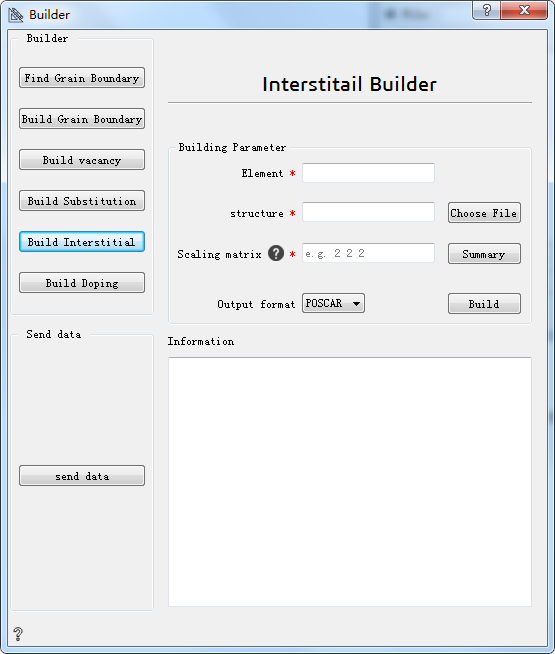
Build Interstitution:build interstitial atom¶
Element* Choose the element to insert interstice
Choose File* Choose the initial structure
Scaling matrix* Scaling matrix used to build supercells. The options include: one integer, representing expand multiple; three vectors,representing three directions at the same time to enlarge the corresponding multiples; nine matrices,representing to enlarge corresponding multiple in matrix.
Summary Offer all the information necessary to build interstitution structure
Output format Choose output format
Build Build interstitial atom
Information Show information about structure of the interstitial atoms
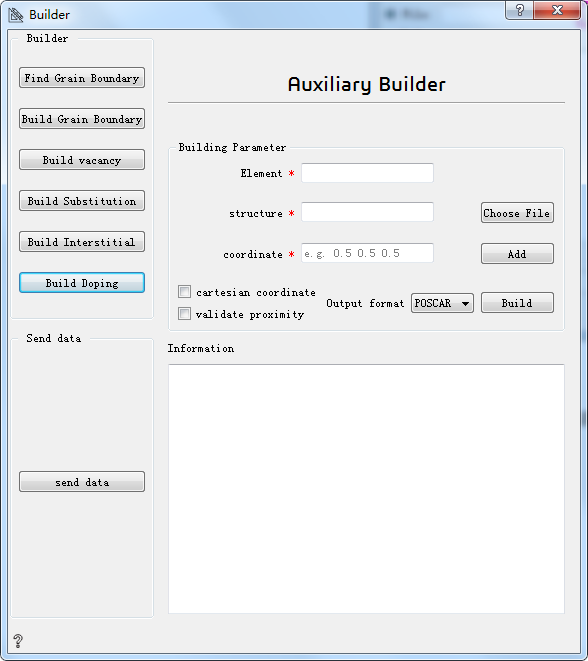
Build Doping:build doping¶
Element* Choose the element to be doped
Choose File* Choose the initial structure
Add* Choose the coordinates of the doped atom. Default is fractional coordinates.
cartesian coordinate Checking indicates that Cartesian coordinates are selected.
validate proximity Checking indicates whether the doping atom position overlaps with the existing atom.
Output format Choose output format
Build Build doping
Information Show information about the structure of doped atoms
Send data:pass the constructed structure information to the next control¶
Multi POSCAR Controls¶
Read multiple files in POSCAR、cif、xsf and xyz formats.
Input |
Output |
|
|---|---|---|
None |
Structure: pymatgen class |
Many Structures |
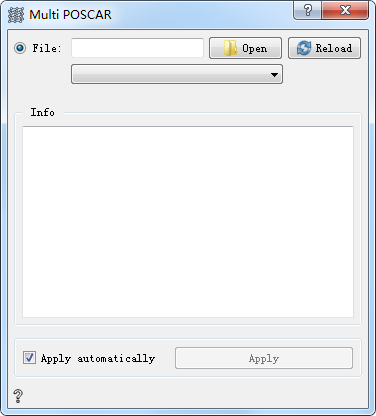
Parameter Configuration:mainly used to load and display atomic structure information, double-click the icon to open the parameter configuration.
Open Open and load multiple files.Drop-down box to select the specified POSCAR file to view.
Reload Reload files
Info Show atomic structure information
Apply automatically If checked, the output of the control can be automatically passed to the next control. If unchecked, you can manually click Apply for transmission, which is checked by default.
Finder Controls¶
Find all possible structures for the specified element in database and generate the corresponding structure file.
Input |
Output |
|---|---|
None |
Data |
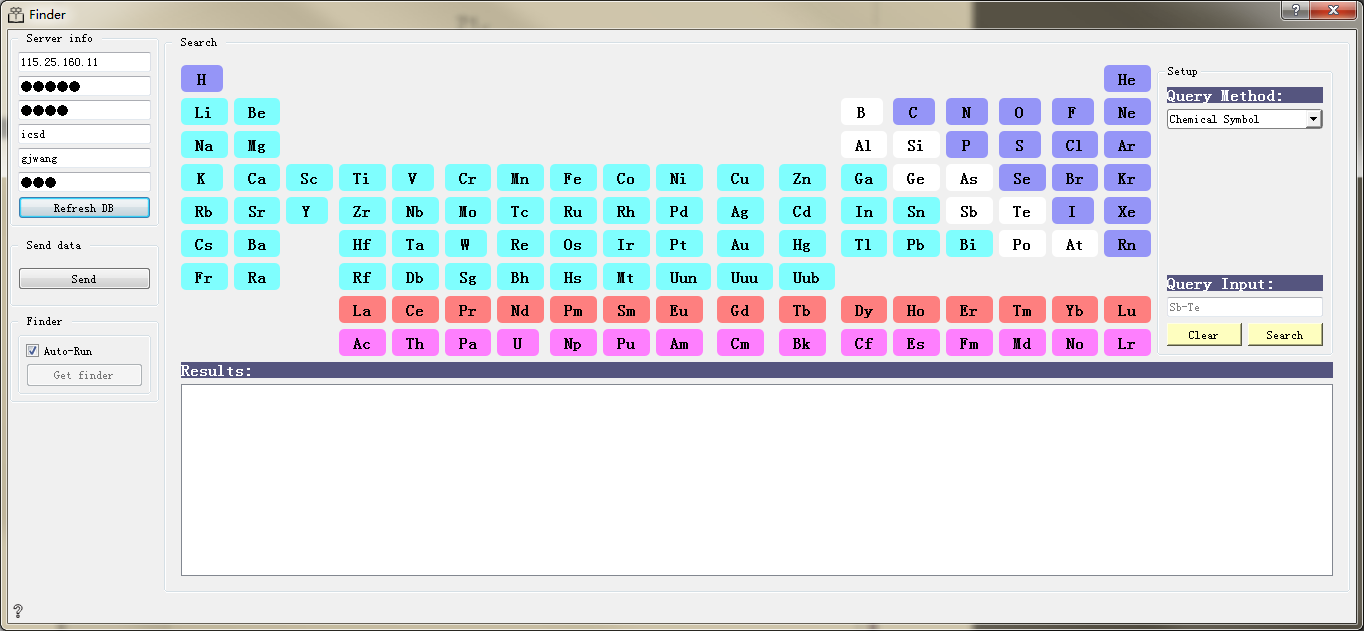
Sever info(Server information):
Sever The server address to connect
Port Server port number
Database Database
collection Collection of MongoDB
Username The user name
Password Password
Refresh DB Refresh database
Structures path The path to local structure files
Send data Send the above data
Search(Find the structure needed):
Click elements in periodic table to find the structure that contains or does not contain the corresponding element, controlled by Setup.
Setup Contains four options, And,And Not,Or,Or Not
Query Method Query methods, including chemical symbols, chemical formulas, regular expressions
Query Input Show the query criteria input
Clear Clear all query conditions
Search Find structures that satisfy the condition
Results Show information of all structures that meet the criteria. Double-click the results you want to view, and can be viewed in visualization software such as Vesta
Finder(send data):
Auto-run If checked, the output of the control can be automatically passed to the next control. If unchecked, you can manually click Get finder for transmission, which is checked by default.
HT Preprocessor Controls¶
High-throughput pretreatment control that is similar to a structural container. All generated structures must be transferred to this control before the next calculation take place.
Input |
Output |
||
|---|---|---|---|
Structure |
Many Structures |
Table |
Data |
Parameter Configuration:
Show the basic information of input structure file
Auto-Run If checked, the output of the control can be automatically passed to the next control. If unchecked, you can manually click Send data for transmission, which is checked by default.
Data Table Controls¶
Display data sets in the form of tables(similar to Excel)
Input |
Output |
|
|---|---|---|
Data |
Data |
Selected Data |
Parameter Configuration:
Info Show information about input data
Variables --> Show variable labels(if present) If checked, variable labels will be showed if present
Variables --> Visualize numeric values Check for visualize numeric values
Variables --> Color by instance classes If checked, the color will be showed by instance classes
Selection --> Select full rows If checked, a whole line will be choosed at a time
Restore Original Order Restore original order
Send Automatically If checked, the output of the control can be automatically passed to the next control. If unchecked, you can manually click Send Automatically for transmission, which is checked by default.
File Controls¶
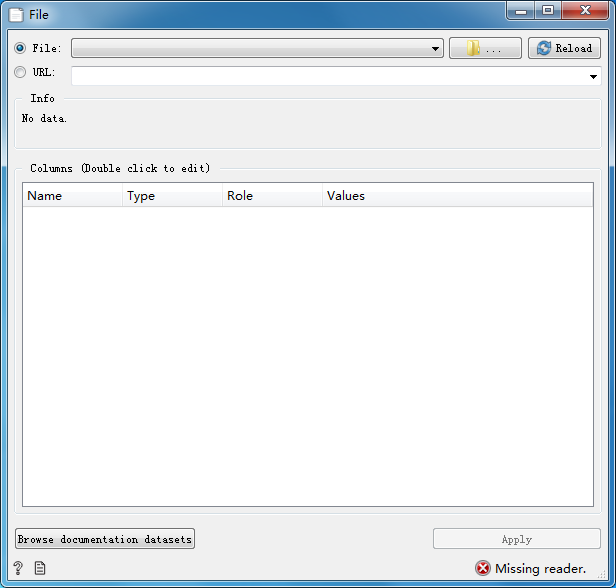
Read data from input file or network and output a data table.
Input |
Output |
|---|---|
None |
Data |
Parameter Configuration:
File Open and load file
Reload Reload file
URL Input the URL of the network file
Info Show information about the file
Columns(Double click to edit) Columns display data information, double-click to edit
Browse documentation datasets Browse documentation datasets
Data Info Controls¶
Show the basic information of the dataset, such as the number and type of variables in each column, how many rows there are, and so on.
Input |
Output |
|---|---|
Data |
None |
Parameter Configuration:
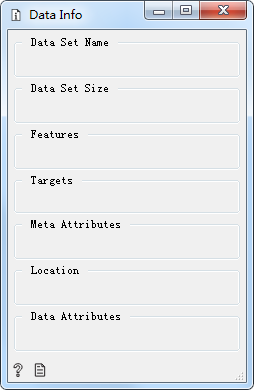
Data Set Name Dataset name
Data Set Size Dataset size
Features Dataset features
Targets
Meta Attributes Meta attributes
Location Dataset storage location
Data Attributes Data attributes
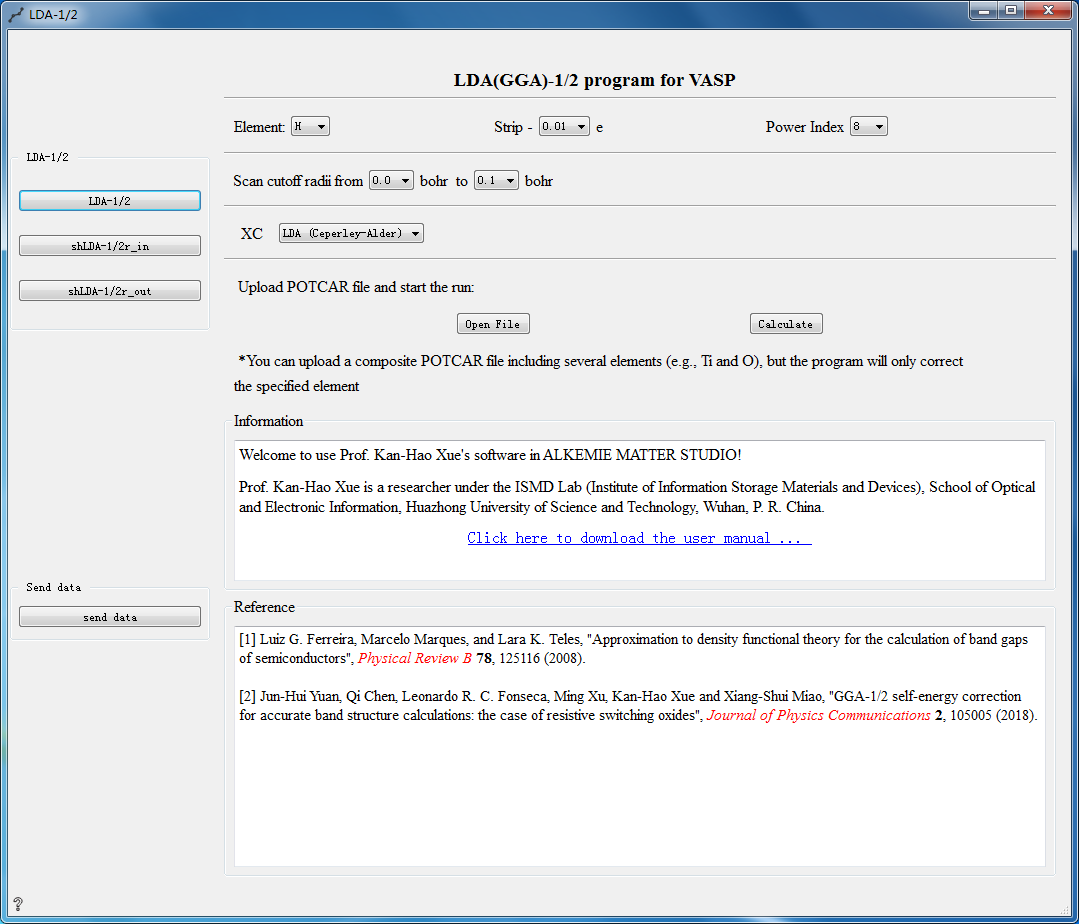
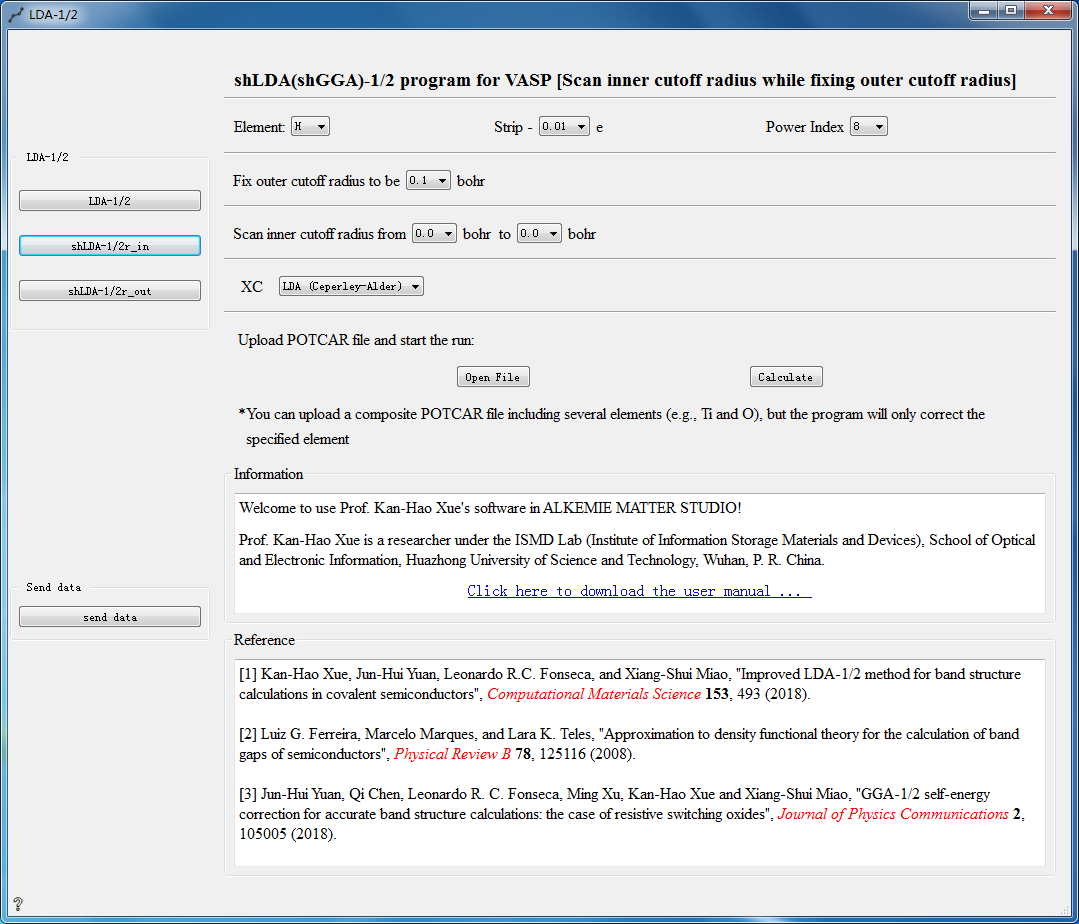
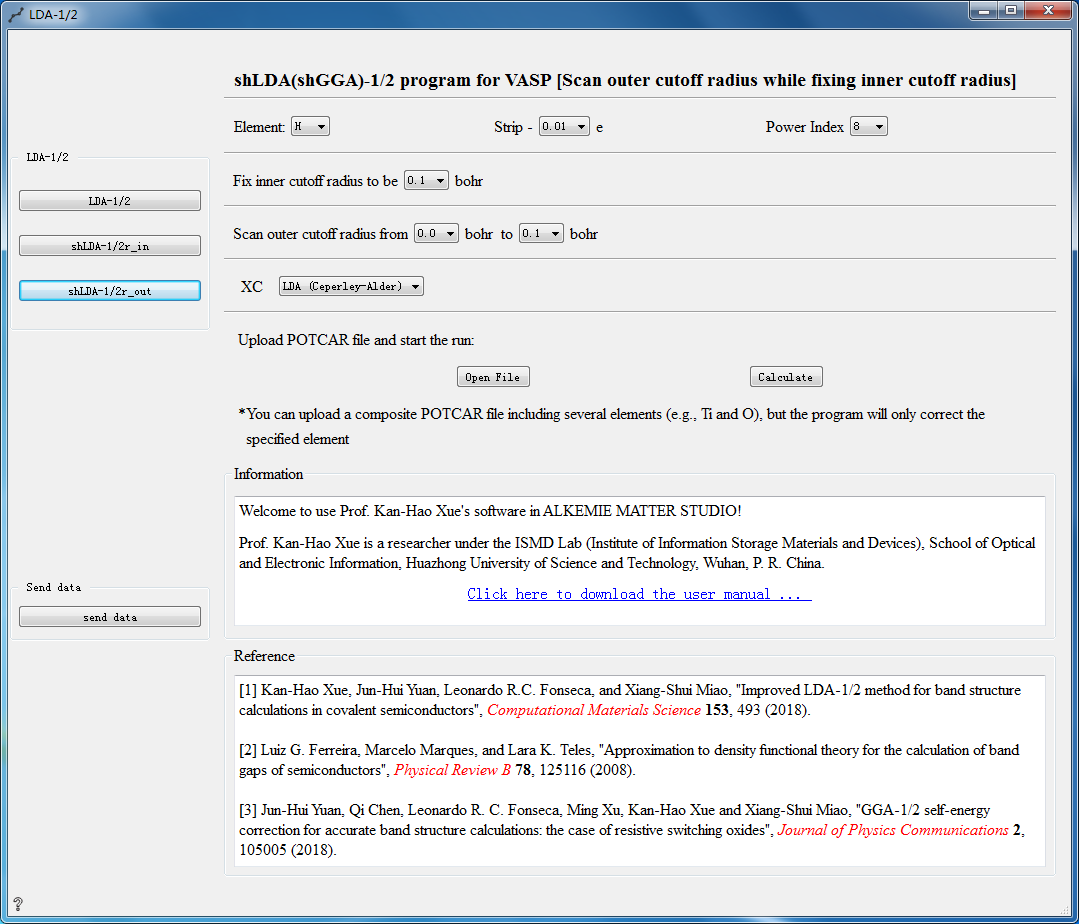
LDA-1/2 Controls¶
Generate special LDA-1/2 potential function
Input |
Output |
|---|---|
file path |
file path |
The following three calculation methods can be choosed for LDA-1/2 calculation:
Parameter Configuration:
Element Choose element
Strip Set self-energy potential modification value
Power Index Set power index
Fix outer cutoff radius Fix outer cutoff radius
Scan inner cutoff radius Scan inner cutoff radius
XC Choosed hybrid functional
Open File Choose to upload the POTCAR file
Calculate Calculate the uploaded POTCAR
Information Related introduction of LDA-1/2 calculation method
Reference Reference for calculation methods
Send data Click Send data to send data when all settings are completed
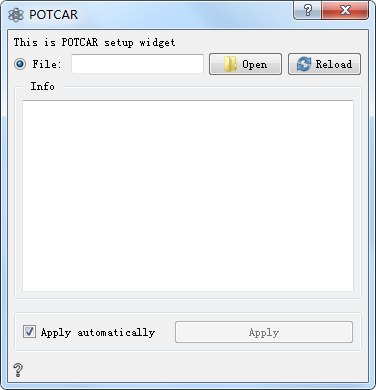
POTCAR Controls¶
Choose the desired POTCAR file
Input |
Output |
|---|---|
file path |
file path |
Parameter Configuration:
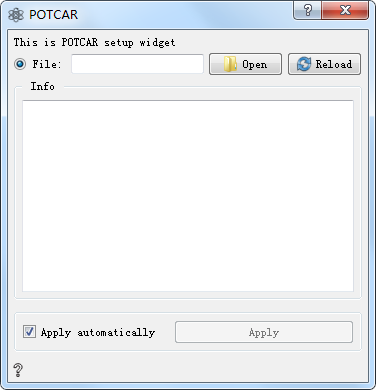
Open Open and load the file
Reload Reload file
Info Show POTCAR file information
Apply automatically If checked, the output of the control can be automatically passed to the next control. If unchecked, you can manually click Apply for transmission, which is checked by default.
1 to 4 Controls¶
Input binary material AB file in cif format, and output AA,AB,BA,BB,4 POSCAR files.
Input |
Output |
|---|---|
file path |
list:many structures |
Parameter Configuration:
Open Open and load the file
Reload Reload file
Info Show information of 4 generated POSCAR files
Apply automatically If checked, the output of the control can be automatically passed to the next control. If unchecked, you can manually click Apply for transmission, which is checked by default.